Adapting efficient multiplexing technologies: Array-in-Well and Mesoscale
Novel Technologies used in HEDIMED
One of the main aims of the HEDIMED project is to investigate what is behind the epidemic of immune-related diseases, and there is a wide range of different exposures under research. Among other things, several infections have been associated with autoimmune diseases or the mechanisms behind them. For example, some enteroviruses have been associated with type 1 diabetes and its preclinical state 1. Childhood infections caused by enteroviruses and adenoviruses have been linked to coeliac disease 2, and common viruses behind respiratory infections such as rhinoviruses and RS-virus have been linked to asthma exacerbations and even disease development 3. Additionally, multiple infections during pregnancy and the first years of life may increase the risk of allergic rhinitis in children 4.
Finding associations between diseases and pathogens
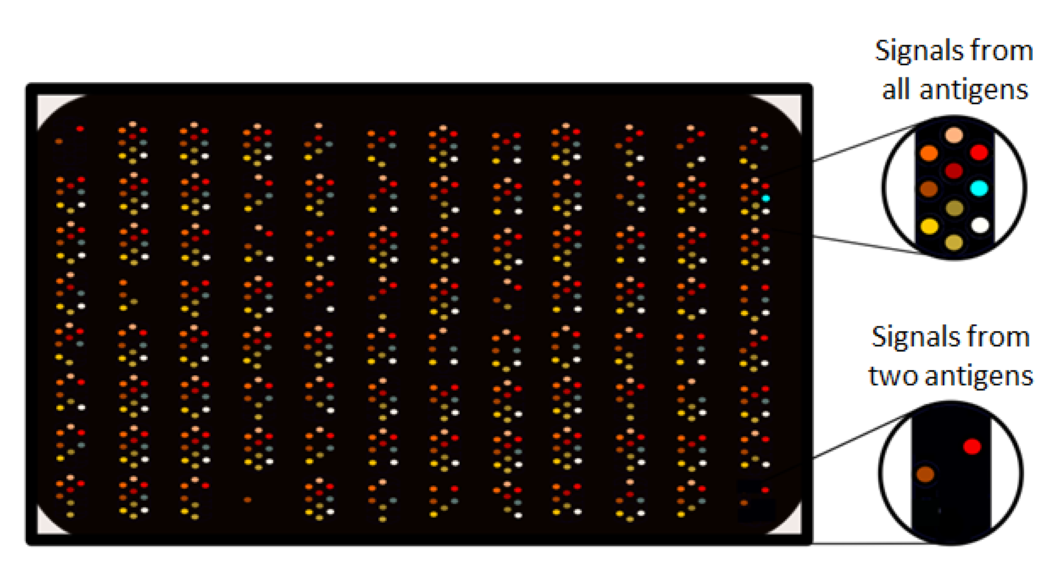
The genetic material of the pathogens doesn’t last in the samples long time, but infecting pathogens leave behind immunological scars (antibodies). These antibodies can be detected with immunological laboratory assays which can detect antibody binding to specific microbial structures (antigens). Depending on the pathogen, such antibodies can be found in blood, stool, urine or saliva. In birth and pregnancy cohorts thousands of blood samples and typically years’ worth of follow-up samples from children and their mothers are collected for future studies. As some of the followed subjects develop a disease at some point, their infection history can be compared to those who didn’t get the disease by analysing antibodies from those early-life samples. See an example in Picture 1. In a nutshell, if the detected antibodies are more prevalent in the group of subjects with the disease, there might be an association. However, showing causality requires much more research. You may read the basis of birth cohorts and how they are utilized in research from a previous HEDIMED blog post.

Need for tools enabling wide-scale research
The multiple birth and pregnancy cohorts in HEDIMED, provide a great opportunity to investigate associations between some common pathogens and type 1 diabetes, celiac disease, allergies and asthma in parallel. Such wide-scale studies have previously been missing, and HEDIMED aims to answer this research need. HEDIMED partners VTT and Tampere University have been adapting more efficient technologies for the analysis of microbial and other antigens in human exposome studies. They aim to compete with and replace conventional assay methods (ELISA/EIA) with more time-, sample-, and material-saving multiplexed methods. These include MesoScale Diagnostics UPLEX platform (MSD) and Array-in-Well (AiW) or a combination of both technologies (Picture 2). AiW is an in-house assay platform of the Technical Research Centre of Finland (VTT). MSD UPLEX is a commercially available platform technology applied by Tampere University for these multiplex microbial antibody analyses.
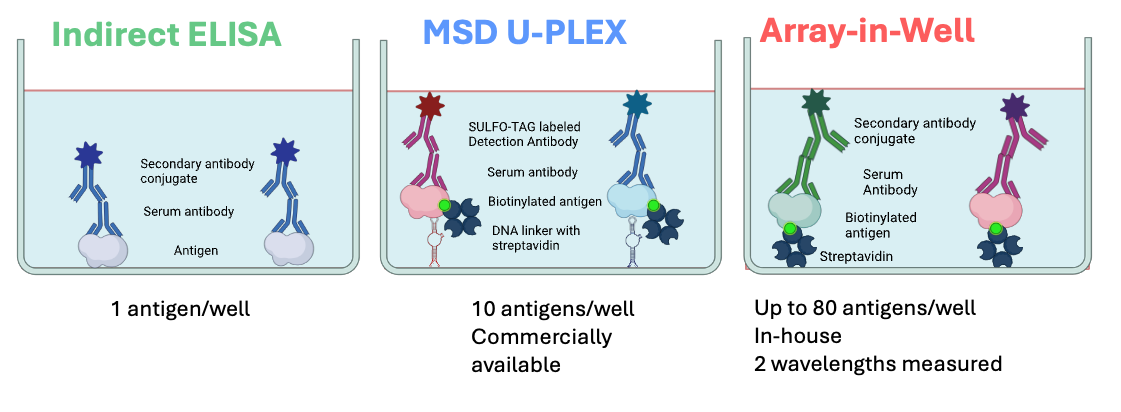
ELISA is still the golden standard when it comes to antibody analyses, but as can be seen in Picture 2, it can detect antibodies against just one microbial antigen at a time. Therefore, studying multiple antigens with ELISA is relatively material (assay plates etc.), sample and time consuming. Both the newer technologies utilized in HEDIMED have a wider dynamic measurement range in comparison to ELISA, and the possibility to detect multiple antigens in parallel. In other words, we need to run fewer dilutions and will get more information from the same volume of blood samples. The technologies differ also from each other, and both have their own advantages. See pictures 3 and 4.

Development of the tools
The first step in developing these tools has been to identify and purchase (or produce) the microbial antigens of interest. The antigens also have to be treated in a certain way (biotinylation) to make them compatible with the multiplex technologies. Following these measures, the researchers started to validate MesoScale and AiW against ELISA/EIA. Simply put, it means that assays have been run with the same microbial antigens with all three technologies and the results have been compared. 10 antigens have been compared with all three technologies, and 40 antigens with the multiplexing technologies. This is often referred to as cross-validation.
The validation process has just been completed and HEDIMED researchers have proceeded to the next step. HEDIMED cohort samples are just beginning to be analysed. Most of the planned 10,000 samples will be analysed with AiW. MSD will be mainly utilized in future studies after HEDIMED for specific sets of antigens, e.g. enteroviruses 5. The advantage of MSD is that it’s commercially available and so accessible for also other study groups and institutions.
Contact
If you want to learn more about the introduced technologies, please contact the following experts:
AiW: Petri Saviranta, VTT, Finland, petri.saviranta@vtt.fi
MesoScale: Niila Jouppila, Tampere University, Finland, niila.jouppila@tuni.fi
Contributions:
Drafting and writing: Minna Turppa and Niila Jouppila
Editing: Niila Jouppila, Heikki Hyöty, Jutta Laiho and Minna Turppa
Pictures: 1, 2 and 4 Niila Jouppila, 3 Petri Saviranta
References:
- Enteroviruses and risk of islet autoimmunity or type 1 diabetes: systematic review and meta-analysis of controlled observational studies detecting viral nucleic acids and proteins. Isaacs, S.R., Roy, A., Dance, B., Ward, E.J., Foskett, D.B., Maxwell, A.J., Rawlinson W.D., Kim, K.W., & Craig, M.E. Lancet Diabetes Endocrinol 2023; 11: 578–92 https://doi.org/10.1016/S2213-8587(23)00122-5
- Review article: exposure to microbes and risk of coeliac disease. Størdal, K., Kahrs, C., Tapia, G., Agardh, D., Kurppa, K. & Stene, L.C. Aliment Pharmacol Ther. 2021, 53:43–62. DOI: 10.1111/apt.16161
- Role of viruses in asthma. Tuomas Jartti, Klaus Bønnelykke, Varpu Elenius & Wojciech Feleszko. Seminars in Immunopathology. 2020, 42:61–74. https://doi.org/10.1007/s00281-020-00781-5
- Early exposure to infections increases the risk of allergic rhinitis-a systematic review and meta-analysis. JunRong, C., Xiaohua, L., Zixin L., Yaqia, Z., Li, X., Jiali, Z., Jin, T., Yide, Y., Mei, T., Yunpen, D. & Jian, L. BCM Pediatrics. 2023 Mar 1;23(1):96. 10.1186/s12887-023-03870-0 https://pubmed.ncbi.nlm.nih.gov/36859178/
- Assessment of Enterovirus Antibodies during Early Childhood Using a Multiplex Immunoassay. Jouppila, N., Lehtonen, J., Seppälä, E., Puustinen, L., Oikarinen, S., Laitinen, O.H., Knip, M., Hyöty, H. & Hytönen V.P. Microbiology Spectrum. 2023, Vol. 1 No. 3. https://doi.org/10.1128/spectrum.05352-22







